
On-the-fly clustering for exascale molecular dynamics simulations.
Alizée Dubois and Thierry Carrard

Summary (AI generated)
In summary, we have developed two algorithms for connected component labeling. The first algorithm, inspired by image processing techniques, is our Formula One implementation. This algorithm is scalable and has been successfully extended to MPI-level scalability. However, optimizing this code requires significant complexity.
The second algorithm is an iterative and alternative approach that is fully scalable, even in worst-case scenarios. From a memory perspective, it maintains scalability and robustness, producing deterministic connected component labeling regardless of the number of MPI processes. This code is straightforward, consisting of only a few lines, and is expected to be portable to GPU architectures in the near future.
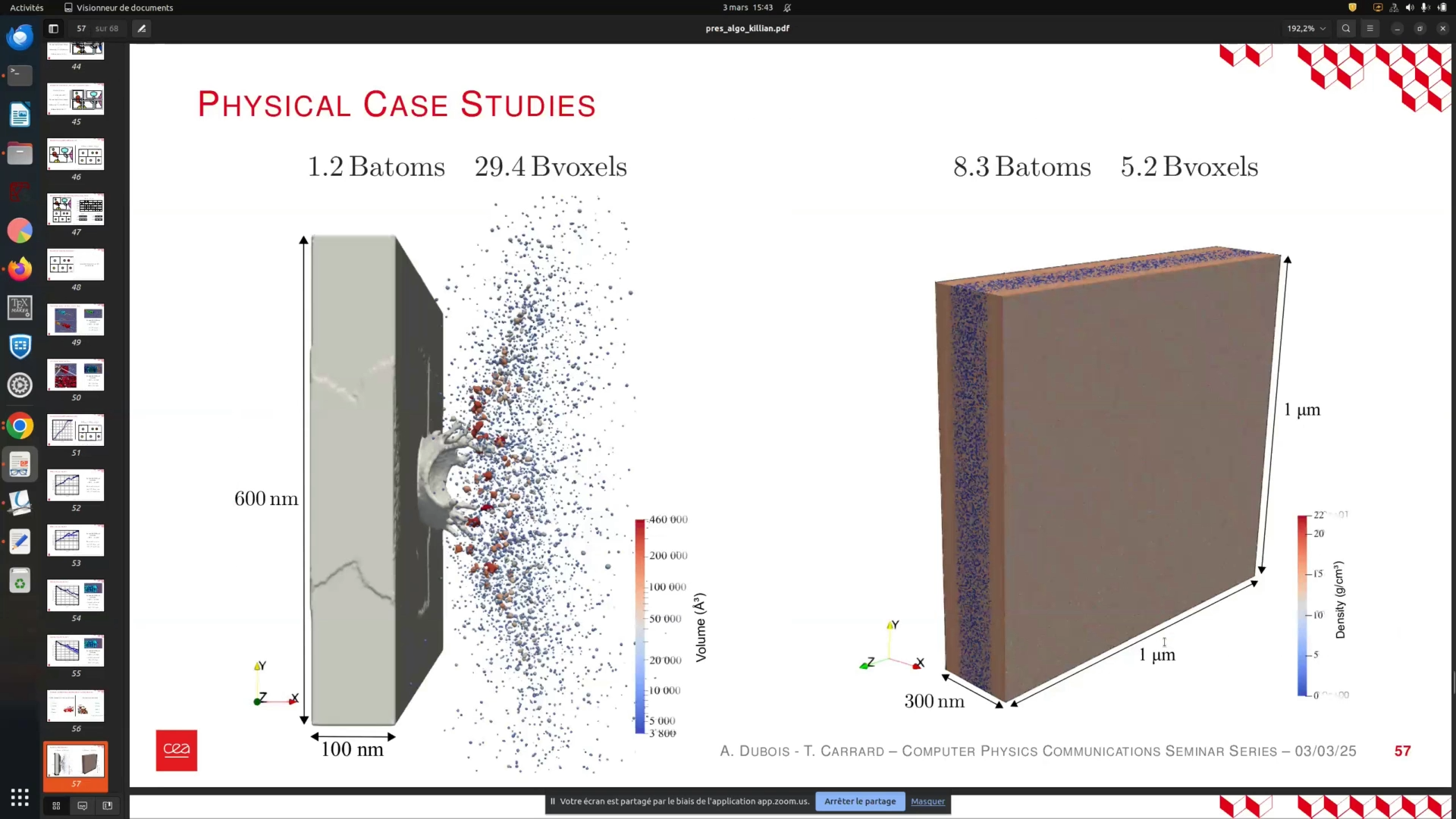
Now, let's explore the applications of these algorithms. The first example illustrates the distribution of mass and velocity of aggregates resulting from liquid impacts on a surface. The second example focuses on ball fracture, where the blue regions represent voids. This allows us to assess whether the simulation is sufficiently large to capture the microscopic properties of small fractures. The accompanying diagram depicts the correlation between compressive wave reflection, tension inflation of voids, velocity at the rear of the sample, and the number of void nuclei.