On-the-fly clustering for exascale molecular dynamics simulations.
Alizée Dubois and Thierry Carrard

Summary (AI generated)
The image processing method for developing connected components is based on the concept of image connected components. We will now shift our focus to a new implementation from a graph processing perspective. As previously mentioned, the algorithm developed during the PhD is efficient and highly optimized. However, it has the limitation of aggregating condensed information to a central MPI node.
To address this, we aimed to create an alternative algorithm for two primary reasons. First, we sought a simpler algorithm that could be easily adapted for GPU implementation in the near future. Second, in cases involving large simulations, we wanted to ensure robustness, avoiding any implementation that centralizes all information on a single node. The alternative implementation has recently been finalized, and I will present it to you now.
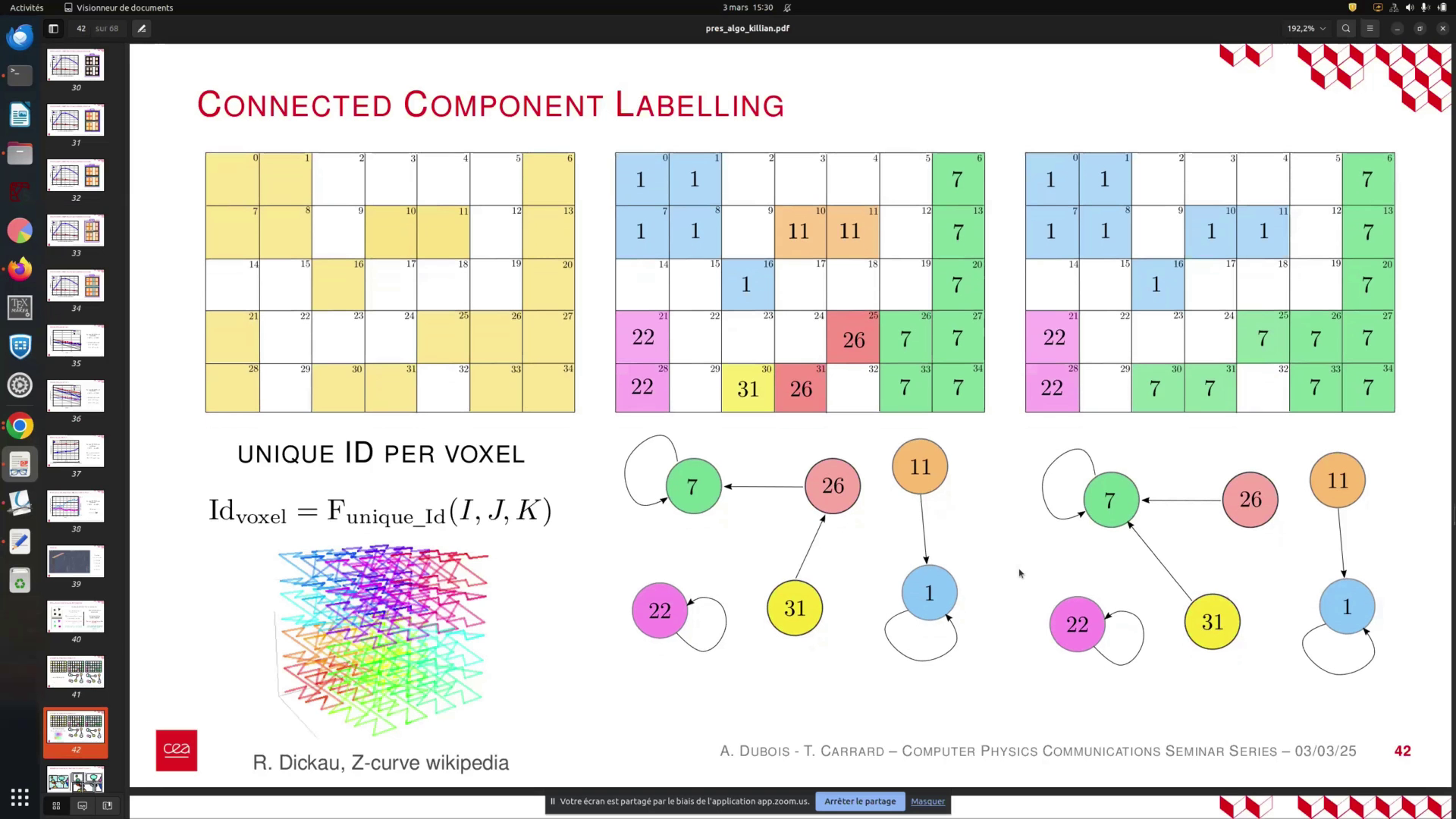
The starting point remains the same as before. From the perspective of the local MPI process, our approach is similar to that of the previous algorithm. The key difference lies in the function used to associate a unique Voxel ID with the coordinates of each cell. This function is crucial to the algorithm's effectiveness and must be deterministic, invariant to the number of MPI processes, and exhibit locality features, which is why we employ a curve-based function.
Additionally, it is essential to know the main and maximum possible values for this function in advance, prior to propagating the labels. These properties are significant, and their importance will become clear shortly. We begin with unique IDs assigned to each voxel and then propagate by identifying the minimum label ID within the connected components.