Preamble
lecture is The Human Virome in Health and Disease.
I will begin by presenting an Introduction to the Human Virome. Next, I will discuss a study on the establishment of the Human Virome. If time allows, I will explore the interrogation of metagenomic dark matter for new viruses, focusing on the search for new viruses associated with humans.
The planetary virome is vast, with approximately 10^7 viruses per ml in seawater. These numbers are astounding, as viruses outnumber their hosts by a factor of 10. When considering the volumes of the oceans, there are roughly 10^31 viral particles on Earth, making them the most successful biological entities. Below are some images of marine phages from Lita Proctor.
The Human Virome is vast, with many people familiar with persistent and latent infections. More than 50% of all adults are commonly positive for herpes viruses, CMV, EBV, HSV1, HHV6, HHV7, Polyomavirus, and Anelloviruses. Pathogenic viruses like HIV are also common in the human population, with roughly 0.8% of people testing positive. Some viruses, like SARS-CoV-2 and cold viruses, are transient and very common. Vaccinia virus vaccine strain is sometimes deliberately used to prevent smallpox infection.
The human genome contains about 8% recognizable fragments of endogenous retroviruses that infected the germ line in the primate lineage leading to humans. Long terminal repeat (LTR) sequences are common in human DNA, comprising 8% of the genome. Additionally, the human microbiome contains numerous viruses, bacteria, phage of bacteria, and archaea in human stool samples. When virus-like particles are purified and stained under a microscope, around 10 to the ninth virus-like particles per gram of human stool can be observed.
In virome studies, specimens are taken from humans such as stool, tissue, or saliva. The solid waste is spun down and filtered at 0.2 um to remove cells. For further purification, a density gradient and chloroform extraction can be done. Nuclease can be used to remove unwanted material, followed by purification, breaking open particles, and purifying nucleic acids (DNA and RNA) before sequencing to obtain a sample of viral population based on sequence information.
A favorable preparation from gut A shows numerous particles, resembling stars in the sky, after fluorescent staining.
We are beginning to understand which viruses are found in different parts of the human body. The gut and oral viral populations are the most numerous, with large numbers of phages being the most common. Bacterial viruses appear to be the most abundant. Animal cell viruses are also present but are usually outnumbered by phages. Despite this, many reads generated from sequence surveys do not align with anything in databases. This is likely due to the vast number of viral populations on Earth, with only a small fraction being sequenced and present in databases. Additionally, viruses evolve quickly, so it is not surprising that most sequences do not have a strong match in a database.
Factors such as the cellular microbiome, diet, genetics, aging, disease, geography, cohabitation, medication, and breastfeeding can all affect viral populations. These factors impact the microbial host and can also have other effects. I will discuss some of these factors further as we continue.
Viruses can influence human health in various ways. Some viruses directly cause pathogenic infections, while others can trigger a protective immune response that may prevent infection by other pathogens. Viruses can activate immune responses through TLR signaling and other molecules, which can be either beneficial in preventing infection or detrimental if the immune response is ineffective and leads to infection by another pathogen.
In addition, viruses can modulate bacterial abundance as predators and transfer DNA into bacteria, influencing their fitness and virulence. For example, cholera toxin is carried on a phage, so the severity of cholera infection depends on the presence of this phage. Overall, we are just beginning to understand the many ways in which viruses can impact human health.
We conducted a study on the establishment of the human virome. Babies were born, and their very first bowel movement, meconium, was collected. Virus-like particles were purified, stained, and examined under a microscope. However, very little was observed in the samples.
A month later, we observed rich viral populations in the sky. We want to ask where all this virus is coming from. We conducted a study of babies studied longitudinally, and I will share the results with you.
The development of the virome in healthy newborns has been studied by various researchers. Lim et al. found no virome in amniotic fluid in healthy pregnancies. The focus of our discussion will be on the virome in healthy infants, as viral infections can severely impact neonatal health. Breitbart et al. conducted a study on one infant and initially found no particles in meconium, but later observed measurable levels. Other studies have shown that high levels of viral particles can be detected in baby stool within 37 hours to two days after birth. Overall, it appears that there are few to no particles present at birth, with rapid colonization occurring shortly after. The virome remains stable later in life, as supported by several studies on microbial colonization of newborns.
Early studies found it difficult to culture bacteria from placenta, meconium, or amniotic fluid, leading to the belief that the womb was sterile. However, later work using PCR and deep sequencing suggested the possible presence of bacterial DNA in various compartments, including the placenta and fetus. This sparked controversy.
A sampling study conducted by colleagues at Penn, in which we participated, did not find a placenta microbiome above the contamination background. Other studies, including one from the Sanger Center, have also reached this conclusion. Additionally, experiments with germ-free animals, such as mice and humans, support the idea that the womb is sterile and colonization occurs shortly after birth.
I believe that infants are colonized with bacteria after the rupture of membranes and delivery, a concept that will be further explored in the colonization study I will be describing.
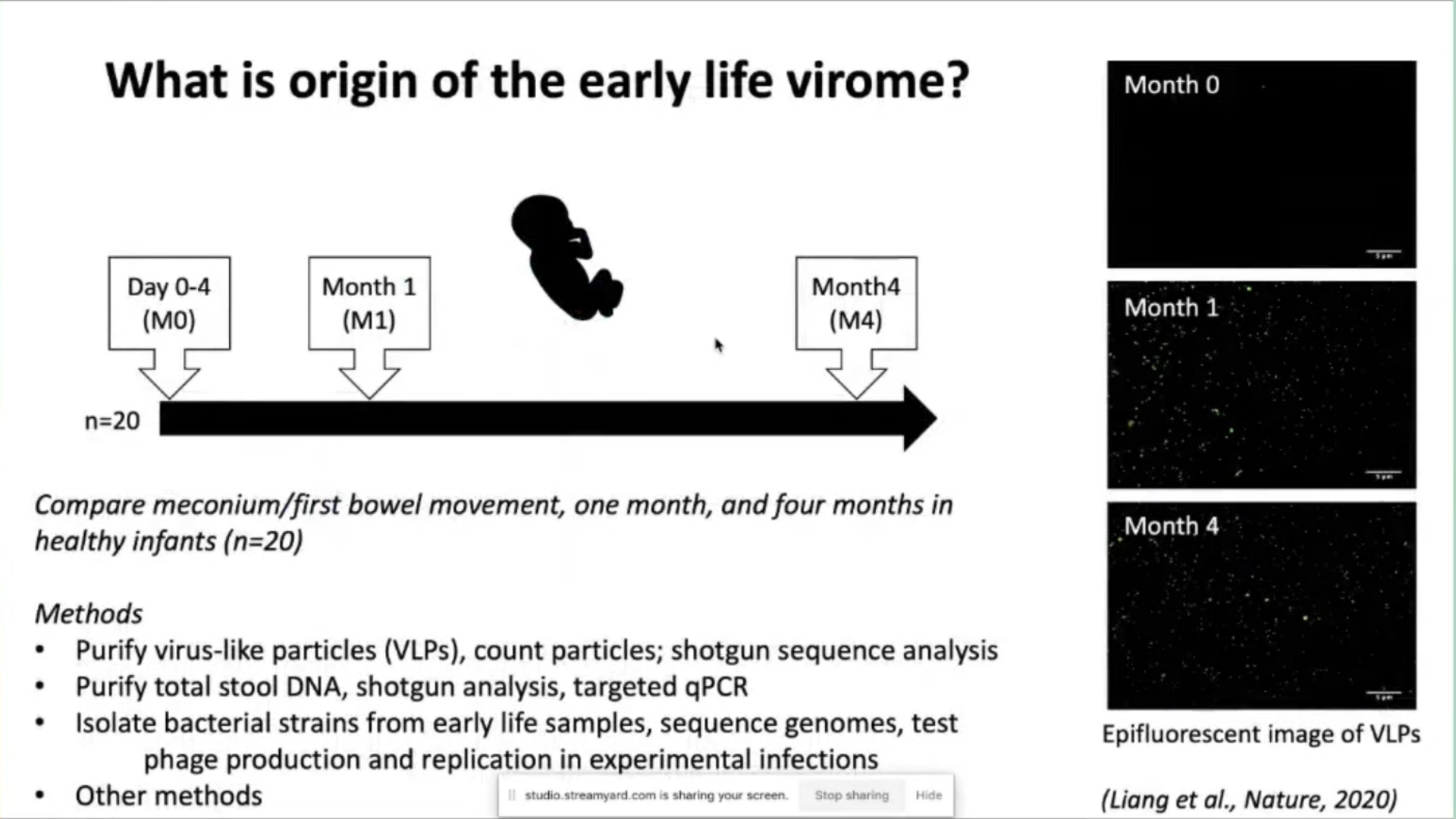
Particles appeared within one month and remained stable. Virus-like particles per gram of feces were low or absent initially but increased over time in the 20 babies studied in our first cohort.
Most babies do not have much 16S rRNA present at birth. Colonization typically occurs soon after birth. This data, along with other information, suggests that the womb is usually sterile when a baby is born and colonization happens quickly.
The origin of the early life virome was investigated through a series of experiments. Virus-like particles were purified and counted, then sequenced. Total stool DNA was also purified and analyzed using shotgun analysis and targeted qPCR. Bacterial strains from early life samples were isolated, their genomes sequenced, and phage production and replication were tested. The results of these studies will be summarized.
In the sequence data, bacteriophages were the most abundant form, while animal cell viruses were rarely observed. Each column represents a sample from a baby, with each row indicating a type of virus - phages or animal cell viruses. By month four, there was a slight increase in the presence of animal cell viruses.
We observed a large number of phage and wondered about their growth. Phage can have two main life cycles: lytic and temperate. In the lytic cycle, phage infect a bacterial cell, replicate, and produce new particles. In the temperate cycle, phage integrate their DNA into a host cell genome and can switch to the lytic cycle under certain conditions.
We were curious about the growth of the phage we observed - whether it was lytic or lysogenic. To investigate, we isolated 24 bacterial strains from the stool of 20 infants and attempted to plate virus-like particles on each strain to look for plaques. Despite our efforts, we did not find any evidence of the infant virus-like particles growing when placed back on cells from the same infant.
This lack of growth led us to consider the possibility of lysogeny, where phage are unable to infect cells that are already lysogens producing repressors. This information suggests that lysogeny may be occurring in this case.
A computational classification of the phage lifestyle using PHACST suggested that phages tend to be more temperate than lytic. This points towards temperate life cycle-induced prophages rather than lytic growth.
We took 24 strains from the babies and tested if they were producing phage. Each row represents one of the 24 strains, with columns showing the conditions used for induction - either with an inducer (mitomycin C) or without, and under aerobic or anaerobic conditions. The number of particles produced is displayed in the table.
It was observed that over half of the strains produced phage, especially in the presence of DNA damage. In fact, 80% of the strains produced significant levels of virus-like particles. This suggests that a large amount of virus is being produced when induced in the presence of DNA damage.
We sequenced the genomes of the bacteria and phage, then compared them. In many cases, we found the phage genomes integrated into the bacteria. For example, in one E. coli strain from a baby, we identified phage genes on the genetic map. When we looked at the strain with or without inducer, we could see the phage present. Similarly, when we examined virus-like particles in the baby's poop, we also found this phage. This suggests that the phage enters the baby through the strain, which harbors a lysogen that the phage induces, leading to a burst and the production of virus-like particles.
While we cannot definitively say this accounts for the entire population, we observed that as the bacteria became more abundant in the baby's poop, the virus-like particles encoded by that bacteria also increased in the stool sample. In some cases, we were able to explain up to half of the viruses present.
We believe that prophage induction from pioneer bacteria is the main mechanism behind the observed early life virome community. Initially, bacteria colonize the baby and commonly have integrated prophage that induce periodically. This results in the first population of viruses that we observe. As time progresses, we may see more lytic phages present in the community.
We observed more animal cell viruses in our study. Upon reviewing our metadata, we discovered a strong association between breastfeeding and a lower recovery of virus-like particles. In our data visualization, yellow represents breastfed babies and green represents formula-fed babies. We identified various viruses in the samples, including adenovirus, anellovirus, astrovirus, calicivirus, enterovirus, and parvovirus. In the first 20 babies, we noticed a higher abundance of viruses in formula-fed babies compared to breastfed babies, although this difference was not statistically significant due to small sample size.
To further investigate, we conducted a study with 125 new babies in the US and found a statistically significant effect of breastfeeding on virus accumulation. We then expanded our research to include African babies, using qPCR for analysis. Our findings in the African samples were consistent with our previous results from the US cohorts. We only had rectal swabs for the African samples, so we used qPCR exclusively for those samples, revealing a significant difference in virus accumulation between breastfed and formula-fed babies.
An effect on phage populations could also be observed. Breastfeeding is known to promote the growth of various bacterial lineages in infants, such as Bifidobacterium and Lactobacillus, along with their associated phages. This suggests that breastfeeding may also have an impact on phages.
In summary, viral colonization in early life appears to occur in a stepwise manner. Healthy babies are typically sterile at birth. The first phase involves the induction of prophage from pioneer bacteria. Subsequently, colonization with viruses infecting human cells occurs, which may be influenced by breastfeeding. The protective effects of breastfeeding have been observed in cohorts from both the US and Africa. Mixed feeding of breast milk and formula has been shown to have a significant suppressive effect on animal cell viruses. This provides encouragement for new mothers to breastfeed as much as possible, even if exclusive breastfeeding is not feasible.
In the last few minutes, I will discuss another study investigating metagenomic dark matter for new viruses.
Some viral infections can be highly pathogenic, while others are common in humans but do not have any known adverse effects, making them appear to be benign commensals. Examples of these viruses include anelloviruses and redondoviruses, which would remain unknown without metagenomic methods. When sequencing, a majority of reads often do not align to anything obvious, leading us to believe they may be common viruses, such as phages, that have not been closely studied before. However, I will now discuss a new family of viruses that we have discovered and find particularly interesting.
We began studying human airway samples during lung transplants. We had two donor-recipient pairs and the students conducting the research noticed a circovirus present. Circoviruses are known as agricultural pathogens and are pathogenic in pigs. This discovery caught our attention. Through database alignment, it was determined that the reads found aligned with a little-known pig virus called porcine stool associated circular virus five. This virus appears to be a CRESS virus, a circular rep encoding single strand DNA virus that has not been extensively studied. When attempting to assemble the reads, the students were able to generate some very detailed genomes.
A redondo virus genome was assembled from two genomes obtained from lung donors. These genomes were used as probes against other respiratory samples, resulting in the discovery of 17 genomes. Another group, Coy et al. in 2017, reported two of these genomes as members of the CRESS virus group. The viruses encode Cp (capsid), Rep (replication initiation) protein, and a third protein of unknown function (ORF3). They were named Redondoviridae, which means "round" in Spanish. The viruses appear to be strictly human-associated, found in respiratory and oral samples.
Further investigation revealed that these viruses were not represented among CRISPR spacers, indicating they are not prokaryotic. Additionally, they do not have prokaryotic Shine-Dalgarno sequences, suggesting they may grow on human cells or eukaryotic organisms associated with humans. Analysis of over 7,581 metagenomic samples confirmed the presence of Redondoviruses, which were found to be associated with periodontitis and critical illness.
In periodontitis samples, redondo virus reads were observed and decreased with treatment in two separate studies.
We conducted our own QPCR studies on critically ill patients in the hospital system at Penn. Our results showed high-level colonization, indicating a cycle of threshold. Low levels were found in the oropharynx or lung of critically ill patients, suggesting long-term persistence related to disease.
Saliva samples from rural populations in Africa were tested and showed an 80% positivity rate. Genome sequencing revealed that both species of redondoviruses are found worldwide. This suggests that redondoviruses have been associated with humans for a long evolutionary period, as both species are widespread.
Respiratory samples from COVID patients were studied, revealing that Redondoviruses are prominent features. A recent paper was published on a study of 83 hospitalized patients and 507 samples, which included targeted qPCR for Anelloviridae and Redondoviridae. It was found that Redondoviridae were significantly higher among intubated patients, while Anelloviridae, considered commensals, were also high. The small circular viruses were the strongest predictor for intubation when attempting to predict the severity of disease, either as intubation or the WHO score in COVID.
A notable predictor for WHO score, we obtained a good area under the curves in each case. There appears to be a connection between these predictors and disease. It is possible that an inflammatory condition is conducive to the growth of these viruses. We are currently investigating the cells on which these viruses grow, with a preference for human cells or human-associated eukaryotes. We are working to gather final data on this topic. Stay tuned for updates.
Summaries of virome studies often reveal sequences that are not found in databases due to the vast numbers of global viruses and their rapid evolution. Some viruses common in humans are benign and only detectable through metagenomic methods, such as Redondoviruses. For example, a study on Redondoviruses began with weak hits in lung transplant samples and has led to further research showing their global ubiquity and enrichment in subjects with periodontitis, intubation, and severe COVID. Researchers are currently searching for the host cell to understand how and where these viruses grow.
In conclusion, this is an introduction to the vast Human Virome. In many studies today, we often encounter things that are completely new to us. The human virome is established by pioneer bacteria with integrated prophages. These bacteria are the first wave of viruses we encounter. We also come across some animal cell viruses, but they are usually suppressed by breastfeeding. Additionally, the discovery of new viruses through metagenomic dark matter is a very interesting and fertile area for exploration.
In conclusion, this is an introduction to the vast Human Virome. In many studies today, the majority of what we observe is new and novel. The human virome includes pioneer bacteria with integrated prophages, which is the first wave. Animal cell viruses are also present but are suppressed by breastfeeding. Searching for new viruses in metagenomic dark matter is a fascinating and promising area for discovery.
Outliine
lecture is The Human Virome in Health and Disease.
I will begin by presenting an Introduction to the Human Virome. Next, I will discuss a study on the establishment of the Human Virome. If time allows, I will explore the interrogation of metagenomic dark matter for new viruses, focusing on the search for new viruses associated with humans.
The planetary virome is vast, with approximately 10^7 viruses per ml in seawater. These numbers are astounding, as viruses outnumber their hosts by a factor of 10. When considering the volumes of the oceans, there are roughly 10^31 viral particles on Earth, making them the most successful biological entities. Below are some images of marine phages from Lita Proctor.
The Human Virome is vast, with many people familiar with persistent and latent infections. More than 50% of all adults are commonly positive for herpes viruses, CMV, EBV, HSV1, HHV6, HHV7, Polyomavirus, and Anelloviruses. Pathogenic viruses like HIV are also common in the human population, with roughly 0.8% of people testing positive. Some viruses, like SARS-CoV-2 and cold viruses, are transient and very common. Vaccinia virus vaccine strain is sometimes deliberately used to prevent smallpox infection.
The human genome contains about 8% recognizable fragments of endogenous retroviruses that infected the germ line in the primate lineage leading to humans. Long terminal repeat (LTR) sequences are common in human DNA, comprising 8% of the genome. Additionally, the human microbiome contains numerous viruses, bacteria, phage of bacteria, and archaea in human stool samples. When virus-like particles are purified and stained under a microscope, around 10 to the ninth virus-like particles per gram of human stool can be observed.
In virome studies, specimens are taken from humans such as stool, tissue, or saliva. The solid waste is spun down and filtered at 0.2 um to remove cells. For further purification, a density gradient and chloroform extraction can be done. Nuclease can be used to remove unwanted material, followed by purification, breaking open particles, and purifying nucleic acids (DNA and RNA) before sequencing to obtain a sample of viral population based on sequence information.
A favorable preparation from gut A shows numerous particles, resembling stars in the sky, after fluorescent staining.
We are beginning to understand which viruses are found in different parts of the human body. The gut and oral viral populations are the most numerous, with large numbers of phages being the most common. Bacterial viruses appear to be the most abundant. Animal cell viruses are also present but are usually outnumbered by phages. Despite this, many reads generated from sequence surveys do not align with anything in databases. This is likely due to the vast number of viral populations on Earth, with only a small fraction being sequenced and present in databases. Additionally, viruses evolve quickly, so it is not surprising that most sequences do not have a strong match in a database.
Factors such as the cellular microbiome, diet, genetics, aging, disease, geography, cohabitation, medication, and breastfeeding can all affect viral populations. These factors impact the microbial host and can also have other effects. I will discuss some of these factors further as we continue.
Viruses can influence human health in various ways. Some viruses directly cause pathogenic infections, while others can trigger a protective immune response that may prevent infection by other pathogens. Viruses can activate immune responses through TLR signaling and other molecules, which can be either beneficial in preventing infection or detrimental if the immune response is ineffective and leads to infection by another pathogen.
In addition, viruses can modulate bacterial abundance as predators and transfer DNA into bacteria, influencing their fitness and virulence. For example, cholera toxin is carried on a phage, so the severity of cholera infection depends on the presence of this phage. Overall, we are just beginning to understand the many ways in which viruses can impact human health.
We conducted a study on the establishment of the human virome. Babies were born, and their very first bowel movement, meconium, was collected. Virus-like particles were purified, stained, and examined under a microscope. However, very little was observed in the samples.
A month later, we observed rich viral populations in the sky. We want to ask where all this virus is coming from. We conducted a study of babies studied longitudinally, and I will share the results with you.
The development of the virome in healthy newborns has been studied by various researchers. Lim et al. found no virome in amniotic fluid in healthy pregnancies. The focus of our discussion will be on the virome in healthy infants, as viral infections can severely impact neonatal health. Breitbart et al. conducted a study on one infant and initially found no particles in meconium, but later observed measurable levels. Other studies have shown that high levels of viral particles can be detected in baby stool within 37 hours to two days after birth. Overall, it appears that there are few to no particles present at birth, with rapid colonization occurring shortly after. The virome remains stable later in life, as supported by several studies on microbial colonization of newborns.
Early studies found it difficult to culture bacteria from placenta, meconium, or amniotic fluid, leading to the belief that the womb was sterile. However, later work using PCR and deep sequencing suggested the possible presence of bacterial DNA in various compartments, including the placenta and fetus. This sparked controversy.
A sampling study conducted by colleagues at Penn, in which we participated, did not find a placenta microbiome above the contamination background. Other studies, including one from the Sanger Center, have also reached this conclusion. Additionally, experiments with germ-free animals, such as mice and humans, support the idea that the womb is sterile and colonization occurs shortly after birth.
I believe that infants are colonized with bacteria after the rupture of membranes and delivery, a concept that will be further explored in the colonization study I will be describing.
Particles appeared within one month and remained stable. Virus-like particles per gram of feces were low or absent initially but increased over time in the 20 babies studied in our first cohort.
Most babies do not have much 16S rRNA present at birth. Colonization typically occurs soon after birth. This data, along with other information, suggests that the womb is usually sterile when a baby is born and colonization happens quickly.
The origin of the early life virome was investigated through a series of experiments. Virus-like particles were purified and counted, then sequenced. Total stool DNA was also purified and analyzed using shotgun analysis and targeted qPCR. Bacterial strains from early life samples were isolated, their genomes sequenced, and phage production and replication were tested. The results of these studies will be summarized.
In the sequence data, bacteriophages were the most abundant form, while animal cell viruses were rarely observed. Each column represents a sample from a baby, with each row indicating a type of virus - phages or animal cell viruses. By month four, there was a slight increase in the presence of animal cell viruses.
We observed a large number of phage and wondered about their growth. Phage can have two main life cycles: lytic and temperate. In the lytic cycle, phage infect a bacterial cell, replicate, and produce new particles. In the temperate cycle, phage integrate their DNA into a host cell genome and can switch to the lytic cycle under certain conditions.
We were curious about the growth of the phage we observed - whether it was lytic or lysogenic. To investigate, we isolated 24 bacterial strains from the stool of 20 infants and attempted to plate virus-like particles on each strain to look for plaques. Despite our efforts, we did not find any evidence of the infant virus-like particles growing when placed back on cells from the same infant.
This lack of growth led us to consider the possibility of lysogeny, where phage are unable to infect cells that are already lysogens producing repressors. This information suggests that lysogeny may be occurring in this case.
A computational classification of the phage lifestyle using PHACST suggested that phages tend to be more temperate than lytic. This points towards temperate life cycle-induced prophages rather than lytic growth.
We took 24 strains from the babies and tested if they were producing phage. Each row represents one of the 24 strains, with columns showing the conditions used for induction - either with an inducer (mitomycin C) or without, and under aerobic or anaerobic conditions. The number of particles produced is displayed in the table.
It was observed that over half of the strains produced phage, especially in the presence of DNA damage. In fact, 80% of the strains produced significant levels of virus-like particles. This suggests that a large amount of virus is being produced when induced in the presence of DNA damage.
We sequenced the genomes of the bacteria and phage, then compared them. In many cases, we found the phage genomes integrated into the bacteria. For example, in one E. coli strain from a baby, we identified phage genes on the genetic map. When we looked at the strain with or without inducer, we could see the phage present. Similarly, when we examined virus-like particles in the baby's poop, we also found this phage. This suggests that the phage enters the baby through the strain, which harbors a lysogen that the phage induces, leading to a burst and the production of virus-like particles.
While we cannot definitively say this accounts for the entire population, we observed that as the bacteria became more abundant in the baby's poop, the virus-like particles encoded by that bacteria also increased in the stool sample. In some cases, we were able to explain up to half of the viruses present.
We believe that prophage induction from pioneer bacteria is the main mechanism behind the observed early life virome community. Initially, bacteria colonize the baby and commonly have integrated prophage that induce periodically. This results in the first population of viruses that we observe. As time progresses, we may see more lytic phages present in the community.
We observed more animal cell viruses in our study. Upon reviewing our metadata, we discovered a strong association between breastfeeding and a lower recovery of virus-like particles. In our data visualization, yellow represents breastfed babies and green represents formula-fed babies. We identified various viruses in the samples, including adenovirus, anellovirus, astrovirus, calicivirus, enterovirus, and parvovirus. In the first 20 babies, we noticed a higher abundance of viruses in formula-fed babies compared to breastfed babies, although this difference was not statistically significant due to small sample size.
To further investigate, we conducted a study with 125 new babies in the US and found a statistically significant effect of breastfeeding on virus accumulation. We then expanded our research to include African babies, using qPCR for analysis. Our findings in the African samples were consistent with our previous results from the US cohorts. We only had rectal swabs for the African samples, so we used qPCR exclusively for those samples, revealing a significant difference in virus accumulation between breastfed and formula-fed babies.
An effect on phage populations could also be observed. Breastfeeding is known to promote the growth of various bacterial lineages in infants, such as Bifidobacterium and Lactobacillus, along with their associated phages. This suggests that breastfeeding may also have an impact on phages.
In summary, viral colonization in early life appears to occur in a stepwise manner. Healthy babies are typically sterile at birth. The first phase involves the induction of prophage from pioneer bacteria. Subsequently, colonization with viruses infecting human cells occurs, which may be influenced by breastfeeding. The protective effects of breastfeeding have been observed in cohorts from both the US and Africa. Mixed feeding of breast milk and formula has been shown to have a significant suppressive effect on animal cell viruses. This provides encouragement for new mothers to breastfeed as much as possible, even if exclusive breastfeeding is not feasible.
In the last few minutes, I will discuss another study investigating metagenomic dark matter for new viruses.
Some viral infections can be highly pathogenic, while others are common in humans but do not have any known adverse effects, making them appear to be benign commensals. Examples of these viruses include anelloviruses and redondoviruses, which would remain unknown without metagenomic methods. When sequencing, a majority of reads often do not align to anything obvious, leading us to believe they may be common viruses, such as phages, that have not been closely studied before. However, I will now discuss a new family of viruses that we have discovered and find particularly interesting.
We began studying human airway samples during lung transplants. We had two donor-recipient pairs and the students conducting the research noticed a circovirus present. Circoviruses are known as agricultural pathogens and are pathogenic in pigs. This discovery caught our attention. Through database alignment, it was determined that the reads found aligned with a little-known pig virus called porcine stool associated circular virus five. This virus appears to be a CRESS virus, a circular rep encoding single strand DNA virus that has not been extensively studied. When attempting to assemble the reads, the students were able to generate some very detailed genomes.
A redondo virus genome was assembled from two genomes obtained from lung donors. These genomes were used as probes against other respiratory samples, resulting in the discovery of 17 genomes. Another group, Coy et al. in 2017, reported two of these genomes as members of the CRESS virus group. The viruses encode Cp (capsid), Rep (replication initiation) protein, and a third protein of unknown function (ORF3). They were named Redondoviridae, which means "round" in Spanish. The viruses appear to be strictly human-associated, found in respiratory and oral samples.
Further investigation revealed that these viruses were not represented among CRISPR spacers, indicating they are not prokaryotic. Additionally, they do not have prokaryotic Shine-Dalgarno sequences, suggesting they may grow on human cells or eukaryotic organisms associated with humans. Analysis of over 7,581 metagenomic samples confirmed the presence of Redondoviruses, which were found to be associated with periodontitis and critical illness.
In periodontitis samples, redondo virus reads were observed and decreased with treatment in two separate studies.
We conducted our own QPCR studies on critically ill patients in the hospital system at Penn. Our results showed high-level colonization, indicating a cycle of threshold. Low levels were found in the oropharynx or lung of critically ill patients, suggesting long-term persistence related to disease.
Saliva samples from rural populations in Africa were tested and showed an 80% positivity rate. Genome sequencing revealed that both species of redondoviruses are found worldwide. This suggests that redondoviruses have been associated with humans for a long evolutionary period, as both species are widespread.
Respiratory samples from COVID patients were studied, revealing that Redondoviruses are prominent features. A recent paper was published on a study of 83 hospitalized patients and 507 samples, which included targeted qPCR for Anelloviridae and Redondoviridae. It was found that Redondoviridae were significantly higher among intubated patients, while Anelloviridae, considered commensals, were also high. The small circular viruses were the strongest predictor for intubation when attempting to predict the severity of disease, either as intubation or the WHO score in COVID.
A notable predictor for WHO score, we obtained a good area under the curves in each case. There appears to be a connection between these predictors and disease. It is possible that an inflammatory condition is conducive to the growth of these viruses. We are currently investigating the cells on which these viruses grow, with a preference for human cells or human-associated eukaryotes. We are working to gather final data on this topic. Stay tuned for updates.
Summaries of virome studies often reveal sequences that are not found in databases due to the vast numbers of global viruses and their rapid evolution. Some viruses common in humans are benign and only detectable through metagenomic methods, such as Redondoviruses. For example, a study on Redondoviruses began with weak hits in lung transplant samples and has led to further research showing their global ubiquity and enrichment in subjects with periodontitis, intubation, and severe COVID. Researchers are currently searching for the host cell to understand how and where these viruses grow.
In conclusion, this is an introduction to the vast Human Virome. In many studies today, we often encounter things that are completely new to us. The human virome is established by pioneer bacteria with integrated prophages. These bacteria are the first wave of viruses we encounter. We also come across some animal cell viruses, but they are usually suppressed by breastfeeding. Additionally, the discovery of new viruses through metagenomic dark matter is a very interesting and fertile area for exploration.
In conclusion, this is an introduction to the vast Human Virome. In many studies today, the majority of what we observe is new and novel. The human virome includes pioneer bacteria with integrated prophages, which is the first wave. Animal cell viruses are also present but are suppressed by breastfeeding. Searching for new viruses in metagenomic dark matter is a fascinating and promising area for discovery.